📝 Emulating Semgrep SAST Pro Taint Mode with Join Mode#
Background#
Semgrep’s Join Mode[1] is a seldom discussed yet interesting experimental feature of the Semgrep OSS SAST engine which can be used to achieve rudimentary taint interprocedural and interfile analyses. In this snippet I document how to combine one search mode and two join mode rules in order to identify intrafile, intraprocedural, interfile and interprocedural tainted sinks.
This is not intended as a tutorial, but I’ll hopefully get one done in the near future as I’ve been deep in the weeds with Semgrep. Having said that, I’ll be brief in an attempt to abide by the Diátaxis[2] information architecture philosophy which underlies https://jdsalaro.com.
📦 All materials relating to this snippet and discussed here are available under jdsalaro/jdsalaro.com
Structure#
As usual with samples and snippets I write, you’ll find the pinned language runtime version within the .tool-versions file. That allows you, or future me, to quickly get the project up and running with 🪄 Install asdf: One Runtime Manager to Rule All Dev Environments.
The only dependency for this snippet is the semgrep Python package[3] and its transitive dependencies. They all can be installed using pipenv sync; as is usual with my Python snippets.
Last, but definitely not least, you’ll find an autogenerated README.md pointing to this blog post and four folders: baseline, join-inline, join-separate, combined.
baseline presents four simple —and hypothetical but realistic— use cases where a Python program processes unsanitized input in an unsafe manner together with a Semgrep rule to flag some of the affected lines.
As is to be expected, baseline misses two crucial scenarios: the interprocedural and interfile ones. Those remaining cases are successfully identified by join-inline. join-separate simply shows how to achieve the same in a more modular way as to be able mix and match rules as necessary. Finally, combined gathers the results of both rule flavors, baseline and join-*, into a single ruleset.
$ tree -L 1
.
├── .tool-versions
├── Pipfile
├── Pipfile.lock
├── README.md
|
├── baseline
├── join-inline
├── join-separate
└── combined
Annotations#
baseline/#
As mentioned previously, within each test case directory we find the same simple Python program src:
baseline
├── rules.yml
├── scan.log
└── src
├── main.py
└── utils.py
main.py contains as the name suggests the __main__ “function” and as well as four different functions vulnerable to injection of myval values due to poor sanitization.
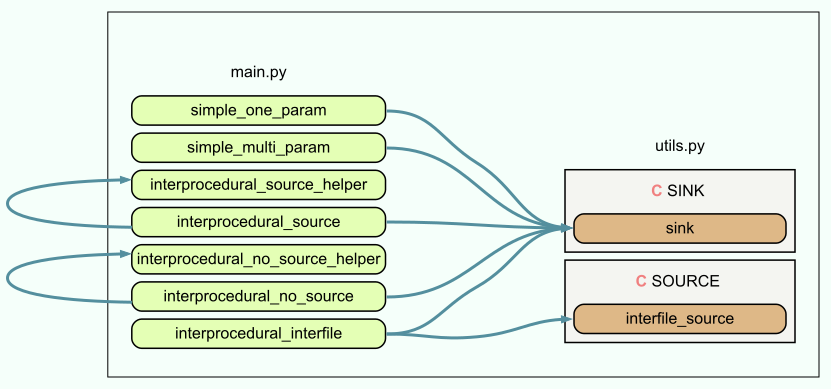
Within main.py and utils.py, the flow of our data of interest RAW pretty much boils down to the following:
It’s worth mentioning that the data flow graph shown is not an textbook UML Data Flow Diagram[4]; a more strictly correct data flow diagram as well as call graph are shown towards the end.
baseline/rules.yml correctly identifies simple_one_param(RAW), simple_multi_param(RAW,RAW) as well as interprocedural_interfile(RAW). It, however, fails to identify cases where RAW is not directly provided and passed through or comes from a function in another file:
1$ semgrep scan -q -c *.yml src > scan.log; cat scan.log
2
3┌─────────────────┐
4│ 3 Code Findings │
5└─────────────────┘
6
7 baseline/src/main.py
8 ❯❯❱ baseline.match
9 match
10
11 20┆ return SINK.sink(tainted)
12 ⋮┆----------------------------------------
13 31┆ return SINK.sink(tainted)
14 ⋮┆----------------------------------------
15 70┆ return SINK.sink(SOURCE.interfile_source(source))
join-inline/#
On the other hand, using join mode, we can detect functions with tainted returns and then separately identify the usage of said tainted returns. Both rules can be put inside a single rules.yml and joined by an on clause such as on: 'first.$FUNCTION < second.$FUNCTION'. Said clause means that second.$FUNCTION must occur within, as in be a substring[5], the first.$FUNCTION call.
1join-inline
2├── rules.yml
3├── scan.log
4└── src
As we can see, this time around all interprocedural and interfile cases where tainted variables cross file or procedure boundaries are identified:
1$ semgrep scan -q -c *.yml src > scan.log; cat scan.log
2
3┌─────────────────┐
4│ 4 Code Findings │
5└─────────────────┘
6
7 baseline/src/main.py
8 ❯❯❱ join-inline.match
9 match
10
11 46┆ return SINK.sink(interprocedural_source_helper(source))
12 ⋮┆----------------------------------------
13 60┆ return SINK.sink(interprocedural_no_source_helper())
14 ⋮┆----------------------------------------
15 70┆ return SINK.sink(SOURCE.interfile_source(source))
16 ⋮┆----------------------------------------
17 75┆ return SINK.sink(SOURCE.interfile_source())
Do realise, however, that the findings initially detected by baseline/rules.yml are missing.
join-separate/#
Through join-separate we can achieve the same as with join-inline, but separate the two rules into different files: tainted-return.yml and
use-of-tainted-return.yml. Then, we can proceed with joining them via rules.yml and the same condition mentioned previously.
join-separate
├── rules.yml
├── scan.log
├── src
├── tainted-return.yml
└── use-of-tainted-return.yml
The findings are, as is expected, equivalent:
1$ semgrep scan -q -c *.yml src > scan.log; cat scan.log
2
3┌─────────────────┐
4│ 4 Code Findings │
5└─────────────────┘
6
7 src/main.py
8 ❯❯❱ match
9 match
10
11 46┆ return SINK.sink(interprocedural_source_helper(source))
12 ⋮┆----------------------------------------
13 60┆ return SINK.sink(interprocedural_no_source_helper())
14 ⋮┆----------------------------------------
15 70┆ return SINK.sink(SOURCE.interfile_source(source))
16 ⋮┆----------------------------------------
17 75┆ return SINK.sink(SOURCE.interfile_source())
combined/#
Finally, with combined we leverage baseline/rules.yml and join-inline/rules.yml to obtain and coalesce the results and thus cover all different injectable sources and sinks found within src/.
combined
├── join-inline.yml
├── rules.yml
├── scan.log
└── src
Note there are duplicate findings, although they can, of course, be trivially removed:
1$ semgrep -q -c rules.yml src> scan.log; cat scan.log
2
3┌─────────────────┐
4│ 7 Code Findings │
5└─────────────────┘
6
7 src/main.py
8 ❯❯❱ match
9 match
10
11 20┆ return SINK.sink(tainted)
12 ⋮┆----------------------------------------
13 31┆ return SINK.sink(tainted)
14 ⋮┆----------------------------------------
15 46┆ return SINK.sink(interprocedural_source_helper(source))
16 ⋮┆----------------------------------------
17 60┆ return SINK.sink(interprocedural_no_source_helper())
18 ⋮┆----------------------------------------
19 70┆ return SINK.sink(SOURCE.interfile_source(source))
20 ⋮┆----------------------------------------
21 70┆ return SINK.sink(SOURCE.interfile_source(source))
22 ⋮┆----------------------------------------
23 75┆ return SINK.sink(SOURCE.interfile_source())
Alternative Visualizations#
Generally speaking, I don’t rely on call graphs and data flow graphs in order to understand how to go about preparing Semgrep rules of low to mid complexity. Nevertheless, they are useful tricks which can help understand more complex scenarios; especially whenever you adventure into less documented areas of the syntax.
CrabViz Call Graph#
CrabViz[6] is a VSCode extension that can be used to generate call graphs which can then be exported to SVG. As far as I’ve observed, they are reliably accurate; probably due to the fact CrabViz uses LSP[7].
UML Data Flow Diagram#
Alternatively, and if we care about a more accurate and useful representation of how tainted data flows through the code, we can create UML Data Flow Diagrams with the Python data-flow-diagram package[8]:
The diagram above is the result of the UML diagram definition found within data-flow-diagram.uml.
Final Words#
That turned out a bit longer than expected but I hope you find these snippets insightful and helpful in your SAST experimentation journey.
Stay in touch by entering your email below and receive updates whenever I post something new:
As always, thank you for reading and remember that feedback is welcome and appreciated; you may contact me via email or social media. Let me know if there's anything else you'd like to know, something you'd like to have corrected, translated, added or clarified further.
